Multimodal Biomarkers for Pregnancy Complications
Verified by Sahaj Satani from ImplementMD



The Implementation Gap
Preeclampsia affects 2–8% of pregnancies globally (70,000 maternal deaths annually), while gestational diabetes mellitus (GDM) complicates 15–20% of pregnancies. Current prediction relies on clinical risk factors achieving only 30–50% sensitivity. Traditional biomarkers (sFlt-1/PlGF) perform well in late pregnancy but lack first-trimester predictive power when intervention is most effective. Breakthrough 2024–2025 validation studies demonstrate cell-free DNA predicts preterm preeclampsia with 81% sensitivity at ≤16 weeks (Adil et al., 2025), cell-free RNA distinguishes disease subtypes 18 weeks before diagnosis (Castillo-Marco et al., 2025), and placental extracellular vesicles identify GDM with 95% sensitivity (Palma et al., 2025). The implementation gap persists: multi-ancestry validation incomplete, clinical workflows undefined, EHR integration pathways unclear, and cost/reimbursement structures unresolved. This brief synthesizes T2 validation evidence and defines T3 implementation readiness criteria.
Evidence for Implementation Readiness
Validated Predictive Models with Clinical Performance
According to PubMed, the PEARL (Preeclampsia Early Assessment of Risk from Liquid biopsy) framework, validated across 1,854 samples at University of Washington/Fred Hutchinson, uses tissue-specific nucleosome profiling from standard NIPT blood draws to predict preterm preeclampsia at ≤16 weeks gestation—12–20 weeks before symptom onset (Adil et al., 2025). Internal validation achieved AUC 0.85 (95% CI: 0.80–0.90) with 81% sensitivity at 80% specificity. External validation confirmed generalizability: AUC 0.84 (0.79–0.89). Early-onset PE (<34 weeks) showed exceptional discrimination: AUC 0.92, 94% sensitivity. Mechanistic signature: decreased placental cfDNA + increased endothelial cfDNA indicates subclinical placental-maternal interface dysfunction.
Spain's PREMOM cfRNA study (NCT04990141), enrolling 9,586 women across 14 hospitals, demonstrated cfRNA enables preeclampsia subtype discrimination months before diagnosis (Castillo-Marco et al., 2025). Early-onset PE (EOPE) prediction at first trimester: AUC 0.88 (internal), 0.87 (external), with 18-week mean lead time. Late-onset PE (LOPE) at second trimester: AUC 0.90 (internal), 0.77 (external), 14.9-week lead time. EOPE signatures reflect decidualization resistance (CBR3, MMP7); LOPE reflects maternal metabolic pathways—enabling personalized intervention (aspirin for EOPE, metabolic management for LOPE).
Table 1. Multimodal Biomarker Platform Performance Metrics
Biomarker Platform | AUC | Sensitivity | Specificity | Lead Time | Key Advantage |
|---|---|---|---|---|---|
cfDNA (PEARL) | 0.85 (0.80–0.90) | 81% | 80% | 12–20 weeks | Bundles with existing NIPT; incremental cost $50–100 |
cfRNA EOPE (T1) | 0.88 (internal) | — | — | 18 weeks | Subtype discrimination enables personalized intervention |
cfRNA LOPE (T2) | 0.90 (internal) | — | — | 14.9 weeks | Maternal metabolic pathway targeting |
Placental EVs (GDM) | — | 95% | 100% | Before 18 weeks | Point-of-care platform; 100% PPV |
sFlt-1/PlGF (Japan) | — | 81.3% | 90% | 1–4 weeks | FDA-cleared; insurance-covered; NPV 98.7% |
Japan's prospective multicenter trial across primary/secondary/tertiary care facilities (n=303) validated sFlt-1/PlGF clinical utility at cut-off 38: NPV 98.7% (95% CI: 96.2–99.3) for ruling out PE within 1 week, PPV 73.7% for adverse outcomes within 4 weeks (Yamazaki et al., 2025). Primary care PPV reached 83.3%, demonstrating usability across all care levels. FDA clearances: Thermo Fisher PreClara (May 2023), Roche Elecsys (February 2025). Japan national insurance coverage since July 2021.
Placental extracellular vesicle nanoplatform for first-trimester GDM prediction achieved 95% sensitivity, 100% specificity, 100% PPV at 11–13 weeks (n=201 women) using CD9+/PLAP+ biomarkers on point-of-care glass strip readout (Palma et al., 2025). Standard fasting glucose achieves only AUC 0.56. Exceptional metrics require independent validation in larger, diverse populations before clinical deployment.
Real-World Implementation and Cost-Effectiveness
PRAECIS US multicenter study (n=1,014; 18 centers; 31% Black, 16% Hispanic) demonstrated sFlt-1/PlGF maintains AUC 0.92 versus 0.75 for standard clinical assessment across racial/ethnic groups. Cost-effectiveness analyses show consistent savings: $10,595/patient (USA), £344/patient (UK), ¥16,373/patient (Japan). PEARL cfDNA projected cost-effective when bundled with NIPT infrastructure—incremental cost $50–100 versus standalone $300–800.
Health Equity Performance Gaps
Critical disparity: Polygenic risk scores show no predictive improvement in women of African ancestry despite improving European-ancestry prediction (Venkatesh et al., 2025). Black women experience 12.4% PE prevalence (versus 7.1% White) and 3.7× increased in-hospital mortality. GDM biomarker scoping review (137 studies, 278 biomarkers) found only 2 studies performed external validation—validated biomarkers remain "scarce and not ready for clinical use" (Rathnayake et al., 2024). PEARL cfDNA and Spain cfRNA require race/ethnicity-stratified validation before broad implementation.
Implementation Solution
Integrated Risk Stratification Algorithm
Sequential biomarker deployment matched to gestational age optimizes sensitivity while controlling costs. The integrated prediction model combines cfDNA/cfRNA for early risk stratification with sFlt-1/PlGF for clinical triage:
First Trimester (9–16 weeks) — Early Risk Stratification:
PEARL model achieves AUC 0.85 when integrating tissue-specific nucleosome patterns with clinical variables. Coefficients (β) derived from XGBoost machine learning. Incremental cost: $50–100 when bundled with existing NIPT infrastructure.
Second Trimester (18–28 weeks) — Subtype Discrimination:
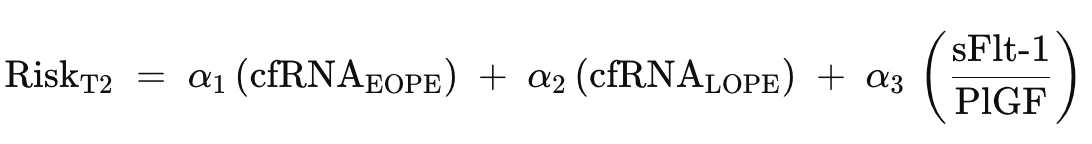
cfRNA enables personalized intervention: EOPE → aspirin + intensive monitoring; LOPE → metabolic management. sFlt-1/PlGF provides established decision support for high-risk patients.
Third Trimester (>28 weeks) — Short-Term Triage:

Clinical Workflow Integration
Deploy via Epic Genomics Module or Cerner Millennium. cfDNA results integrate through existing NIPT reporting infrastructure. sFlt-1/PlGF orders via CPOE with automated Best Practice Alerts for suspected PE. Algorithm-generated risk scores display on chart SnapShot with trimester-specific intervention recommendations.
Table 2. EHR Implementation Timeline
Gestational Age | Biomarker | Action | EHR Integration |
|---|---|---|---|
9–16 weeks | cfDNA + NIPT | Risk stratification: Low/Int/High | Epic Genomics Module auto-populate |
18–28 weeks | cfRNA OR sFlt-1/PlGF (if high-risk) | EOPE: aspirin; LOPE: metabolic mgmt | Best Practice Alert triggers |
>28 weeks | sFlt-1/PlGF (suspected cases) | Discharge (<38) vs Transfer (>85) | Ob Triage decision support |
11–13 weeks (GDM) | Placental EV (if available) | Early lifestyle intervention | Point-of-care result → care plan |
Figure 1: Multimodal Biomarker Clinical Pathway
┌─────────────────────────────────────────────────────────────────────────────┐ │ PREECLAMPSIA & GDM MULTIMODAL BIOMARKER WORKFLOW │ ├─────────────────────────────────────────────────────────────────────────────┤ │ │ │ FIRST TRIMESTER (9-16 weeks) SECOND TRIMESTER (18-28 weeks) │ │ ┌──────────────────┐ ┌───────────────────┐ │ │ │ cfDNA + NIPT │──HIGH RISK──▶ │ cfRNA Subtype OR │ │ │ │ • Nucleosome │ │ sFlt-1/PlGF │ │ │ │ • AUC 0.85 │ │ • EOPE: Aspirin │ │ │ │ • Cost: +$50-100 │ │ • LOPE: Metabolic │ │ │ └────────┬─────────┘ └─────────┬─────────┘ │ │ │ │ │ │ ▼ ▼ │ │ ┌──────────────────┐ ┌───────────────────┐ │ │ │ RISK TIERS │ │ INTERVENTION │ │ │ │ Low: Standard │ │ EOPE → Aspirin + │ │ │ │ Int: Monitor │ │ Monitor │ │ │ │ High → Advanced │ │ LOPE → BP control │ │ │ └──────────────────┘ └───────────────────┘ │ │ │ │ GDM PATHWAY (11-13 weeks) THIRD TRIMESTER (>28 weeks) │ │ ┌──────────────────┐ ┌───────────────────┐ │ │ │ Placental EVs │ │ sFlt-1/PlGF TRIAGE│ │ │ │ • 95% sens │ │ <38 → Discharge │ │ │ │ • 100% spec │ │ (NPV 98.7%) │ │ │ │ • POC platform │ │ >85 → Transfer │ │ │ └────────┬─────────┘ │ (PPV 77.8%) │ │ │ │ └───────────────────┘ │ │ ▼ │ │ ┌──────────────────┐ ┌───────────────────┐ │ │ │ EARLY LIFESTYLE │ │ OUTCOMES │ │ │ │ • Diet + Exercise│ │ • Maternal safety │ │ │ │ • Before OGTT │ │ • Cost savings │ │ │ └──────────────────┘ └───────────────────┘ │ │ │ │ ───────────────────────────────────────────────────────────────────────── │ │ INPUTS: cfDNA nucleosomes • cfRNA transcripts • sFlt-1/PlGF • Placental EVs│ │ METHODS: XGBoost ML • Tissue-specific profiling • Immunoassays │ │ OUTPUT: Risk Score (0-100) + Trimester-specific intervention pathway │ └─────────────────────────────────────────────────────────────────────────────┘
References
Adil, M., Kolarova, T. R., Doebley, A. L., Ha, G., et al. (2025). Preeclampsia risk prediction from prenatal cell-free DNA screening. Nature Medicine, 31(4), 1312–1318. https://doi.org/10.1038/s41591-025-03509-w
Andresen, I. J., Romero, R., Kalapotharakos, G., et al. (2025). Large-scale proteomics reveals new candidate biomarkers for late-onset preeclampsia. Hypertension. https://doi.org/10.1161/HYPERTENSIONAHA.125.25189
Castillo-Marco, N., Cordero, T., Igual, M., et al. (2025). Maternal plasma cell-free RNA as a predictor of early and late-onset preeclampsia throughout pregnancy. Nature Communications, 16, 9208. https://doi.org/10.1038/s41467-025-64215-2
Kelly, C. B., Hookham, M. B., Yu, J. Y., et al. (2018). Subclinical first trimester renal abnormalities are associated with preeclampsia in normoalbuminuric women with type 1 diabetes. Diabetes Care, 41(1), 120–127. https://doi.org/10.2337/dc17-1635
Palma, C., Masud, M. K., Guanzon, D., et al. (2025). Rapid and high-sensitivity screening of pregnancy complications by profiling circulating placental extracellular vesicles. Science Advances, 11(9), eadr4074. https://doi.org/10.1126/sciadv.adr4074
Provendier, A., Azria, E., Kayem, G., et al. (2024). The sFlt-1/PlGF ratio for prediction of preeclampsia-related outcomes in women with preexisting diabetes. Reproductive Sciences, 31, 2371–2378. https://doi.org/10.1007/s43032-024-01540-9
Rathnayake, H., Han, L., da Silva Costa, F., et al. (2024). Advancement in predictive biomarkers for gestational diabetes mellitus diagnosis and related outcomes: A scoping review. BMJ Open, 14(12), e089937. https://doi.org/10.1136/bmjopen-2024-089937
Starodubtseva, N., Tokareva, A., Kononikhin, A., et al. (2025). Proteome-based maternal plasma and serum biomarkers for preeclampsia: A systematic review and meta-analysis. Life, 15(5), 776. https://doi.org/10.3390/life15050776
Venkatesh, K. K., Bello, N. A., Tellez Freitas, C. M., et al. (2025). Polygenic risk scores for preeclampsia prediction beyond gold-standard clinical models in multiethnic populations. Journal of the American Heart Association. https://doi.org/10.1161/JAHA.125.046211
Yamazaki, T., Cerdeira, A. S., Teraoka, Y., et al. (2025). Predictive accuracy of sFlt-1/PlGF ratio for preeclampsia and adverse outcomes: Prospective, multicenter observational study in Japan. Hypertension Research, 48, 2548–2557. https://doi.org/10.1038/s41440-025-02282-0
The Implementation Gap
Preeclampsia affects 2–8% of pregnancies globally (70,000 maternal deaths annually), while gestational diabetes mellitus (GDM) complicates 15–20% of pregnancies. Current prediction relies on clinical risk factors achieving only 30–50% sensitivity. Traditional biomarkers (sFlt-1/PlGF) perform well in late pregnancy but lack first-trimester predictive power when intervention is most effective. Breakthrough 2024–2025 validation studies demonstrate cell-free DNA predicts preterm preeclampsia with 81% sensitivity at ≤16 weeks (Adil et al., 2025), cell-free RNA distinguishes disease subtypes 18 weeks before diagnosis (Castillo-Marco et al., 2025), and placental extracellular vesicles identify GDM with 95% sensitivity (Palma et al., 2025). The implementation gap persists: multi-ancestry validation incomplete, clinical workflows undefined, EHR integration pathways unclear, and cost/reimbursement structures unresolved. This brief synthesizes T2 validation evidence and defines T3 implementation readiness criteria.
Evidence for Implementation Readiness
Validated Predictive Models with Clinical Performance
According to PubMed, the PEARL (Preeclampsia Early Assessment of Risk from Liquid biopsy) framework, validated across 1,854 samples at University of Washington/Fred Hutchinson, uses tissue-specific nucleosome profiling from standard NIPT blood draws to predict preterm preeclampsia at ≤16 weeks gestation—12–20 weeks before symptom onset (Adil et al., 2025). Internal validation achieved AUC 0.85 (95% CI: 0.80–0.90) with 81% sensitivity at 80% specificity. External validation confirmed generalizability: AUC 0.84 (0.79–0.89). Early-onset PE (<34 weeks) showed exceptional discrimination: AUC 0.92, 94% sensitivity. Mechanistic signature: decreased placental cfDNA + increased endothelial cfDNA indicates subclinical placental-maternal interface dysfunction.
Spain's PREMOM cfRNA study (NCT04990141), enrolling 9,586 women across 14 hospitals, demonstrated cfRNA enables preeclampsia subtype discrimination months before diagnosis (Castillo-Marco et al., 2025). Early-onset PE (EOPE) prediction at first trimester: AUC 0.88 (internal), 0.87 (external), with 18-week mean lead time. Late-onset PE (LOPE) at second trimester: AUC 0.90 (internal), 0.77 (external), 14.9-week lead time. EOPE signatures reflect decidualization resistance (CBR3, MMP7); LOPE reflects maternal metabolic pathways—enabling personalized intervention (aspirin for EOPE, metabolic management for LOPE).
Table 1. Multimodal Biomarker Platform Performance Metrics
Biomarker Platform | AUC | Sensitivity | Specificity | Lead Time | Key Advantage |
|---|---|---|---|---|---|
cfDNA (PEARL) | 0.85 (0.80–0.90) | 81% | 80% | 12–20 weeks | Bundles with existing NIPT; incremental cost $50–100 |
cfRNA EOPE (T1) | 0.88 (internal) | — | — | 18 weeks | Subtype discrimination enables personalized intervention |
cfRNA LOPE (T2) | 0.90 (internal) | — | — | 14.9 weeks | Maternal metabolic pathway targeting |
Placental EVs (GDM) | — | 95% | 100% | Before 18 weeks | Point-of-care platform; 100% PPV |
sFlt-1/PlGF (Japan) | — | 81.3% | 90% | 1–4 weeks | FDA-cleared; insurance-covered; NPV 98.7% |
Japan's prospective multicenter trial across primary/secondary/tertiary care facilities (n=303) validated sFlt-1/PlGF clinical utility at cut-off 38: NPV 98.7% (95% CI: 96.2–99.3) for ruling out PE within 1 week, PPV 73.7% for adverse outcomes within 4 weeks (Yamazaki et al., 2025). Primary care PPV reached 83.3%, demonstrating usability across all care levels. FDA clearances: Thermo Fisher PreClara (May 2023), Roche Elecsys (February 2025). Japan national insurance coverage since July 2021.
Placental extracellular vesicle nanoplatform for first-trimester GDM prediction achieved 95% sensitivity, 100% specificity, 100% PPV at 11–13 weeks (n=201 women) using CD9+/PLAP+ biomarkers on point-of-care glass strip readout (Palma et al., 2025). Standard fasting glucose achieves only AUC 0.56. Exceptional metrics require independent validation in larger, diverse populations before clinical deployment.
Real-World Implementation and Cost-Effectiveness
PRAECIS US multicenter study (n=1,014; 18 centers; 31% Black, 16% Hispanic) demonstrated sFlt-1/PlGF maintains AUC 0.92 versus 0.75 for standard clinical assessment across racial/ethnic groups. Cost-effectiveness analyses show consistent savings: $10,595/patient (USA), £344/patient (UK), ¥16,373/patient (Japan). PEARL cfDNA projected cost-effective when bundled with NIPT infrastructure—incremental cost $50–100 versus standalone $300–800.
Health Equity Performance Gaps
Critical disparity: Polygenic risk scores show no predictive improvement in women of African ancestry despite improving European-ancestry prediction (Venkatesh et al., 2025). Black women experience 12.4% PE prevalence (versus 7.1% White) and 3.7× increased in-hospital mortality. GDM biomarker scoping review (137 studies, 278 biomarkers) found only 2 studies performed external validation—validated biomarkers remain "scarce and not ready for clinical use" (Rathnayake et al., 2024). PEARL cfDNA and Spain cfRNA require race/ethnicity-stratified validation before broad implementation.
Implementation Solution
Integrated Risk Stratification Algorithm
Sequential biomarker deployment matched to gestational age optimizes sensitivity while controlling costs. The integrated prediction model combines cfDNA/cfRNA for early risk stratification with sFlt-1/PlGF for clinical triage:
First Trimester (9–16 weeks) — Early Risk Stratification:
PEARL model achieves AUC 0.85 when integrating tissue-specific nucleosome patterns with clinical variables. Coefficients (β) derived from XGBoost machine learning. Incremental cost: $50–100 when bundled with existing NIPT infrastructure.
Second Trimester (18–28 weeks) — Subtype Discrimination:
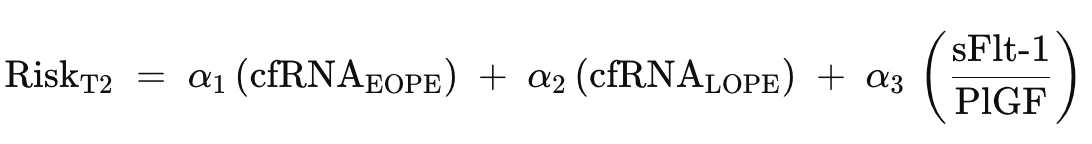
cfRNA enables personalized intervention: EOPE → aspirin + intensive monitoring; LOPE → metabolic management. sFlt-1/PlGF provides established decision support for high-risk patients.
Third Trimester (>28 weeks) — Short-Term Triage:

Clinical Workflow Integration
Deploy via Epic Genomics Module or Cerner Millennium. cfDNA results integrate through existing NIPT reporting infrastructure. sFlt-1/PlGF orders via CPOE with automated Best Practice Alerts for suspected PE. Algorithm-generated risk scores display on chart SnapShot with trimester-specific intervention recommendations.
Table 2. EHR Implementation Timeline
Gestational Age | Biomarker | Action | EHR Integration |
|---|---|---|---|
9–16 weeks | cfDNA + NIPT | Risk stratification: Low/Int/High | Epic Genomics Module auto-populate |
18–28 weeks | cfRNA OR sFlt-1/PlGF (if high-risk) | EOPE: aspirin; LOPE: metabolic mgmt | Best Practice Alert triggers |
>28 weeks | sFlt-1/PlGF (suspected cases) | Discharge (<38) vs Transfer (>85) | Ob Triage decision support |
11–13 weeks (GDM) | Placental EV (if available) | Early lifestyle intervention | Point-of-care result → care plan |
Figure 1: Multimodal Biomarker Clinical Pathway
┌─────────────────────────────────────────────────────────────────────────────┐ │ PREECLAMPSIA & GDM MULTIMODAL BIOMARKER WORKFLOW │ ├─────────────────────────────────────────────────────────────────────────────┤ │ │ │ FIRST TRIMESTER (9-16 weeks) SECOND TRIMESTER (18-28 weeks) │ │ ┌──────────────────┐ ┌───────────────────┐ │ │ │ cfDNA + NIPT │──HIGH RISK──▶ │ cfRNA Subtype OR │ │ │ │ • Nucleosome │ │ sFlt-1/PlGF │ │ │ │ • AUC 0.85 │ │ • EOPE: Aspirin │ │ │ │ • Cost: +$50-100 │ │ • LOPE: Metabolic │ │ │ └────────┬─────────┘ └─────────┬─────────┘ │ │ │ │ │ │ ▼ ▼ │ │ ┌──────────────────┐ ┌───────────────────┐ │ │ │ RISK TIERS │ │ INTERVENTION │ │ │ │ Low: Standard │ │ EOPE → Aspirin + │ │ │ │ Int: Monitor │ │ Monitor │ │ │ │ High → Advanced │ │ LOPE → BP control │ │ │ └──────────────────┘ └───────────────────┘ │ │ │ │ GDM PATHWAY (11-13 weeks) THIRD TRIMESTER (>28 weeks) │ │ ┌──────────────────┐ ┌───────────────────┐ │ │ │ Placental EVs │ │ sFlt-1/PlGF TRIAGE│ │ │ │ • 95% sens │ │ <38 → Discharge │ │ │ │ • 100% spec │ │ (NPV 98.7%) │ │ │ │ • POC platform │ │ >85 → Transfer │ │ │ └────────┬─────────┘ │ (PPV 77.8%) │ │ │ │ └───────────────────┘ │ │ ▼ │ │ ┌──────────────────┐ ┌───────────────────┐ │ │ │ EARLY LIFESTYLE │ │ OUTCOMES │ │ │ │ • Diet + Exercise│ │ • Maternal safety │ │ │ │ • Before OGTT │ │ • Cost savings │ │ │ └──────────────────┘ └───────────────────┘ │ │ │ │ ───────────────────────────────────────────────────────────────────────── │ │ INPUTS: cfDNA nucleosomes • cfRNA transcripts • sFlt-1/PlGF • Placental EVs│ │ METHODS: XGBoost ML • Tissue-specific profiling • Immunoassays │ │ OUTPUT: Risk Score (0-100) + Trimester-specific intervention pathway │ └─────────────────────────────────────────────────────────────────────────────┘
References
Adil, M., Kolarova, T. R., Doebley, A. L., Ha, G., et al. (2025). Preeclampsia risk prediction from prenatal cell-free DNA screening. Nature Medicine, 31(4), 1312–1318. https://doi.org/10.1038/s41591-025-03509-w
Andresen, I. J., Romero, R., Kalapotharakos, G., et al. (2025). Large-scale proteomics reveals new candidate biomarkers for late-onset preeclampsia. Hypertension. https://doi.org/10.1161/HYPERTENSIONAHA.125.25189
Castillo-Marco, N., Cordero, T., Igual, M., et al. (2025). Maternal plasma cell-free RNA as a predictor of early and late-onset preeclampsia throughout pregnancy. Nature Communications, 16, 9208. https://doi.org/10.1038/s41467-025-64215-2
Kelly, C. B., Hookham, M. B., Yu, J. Y., et al. (2018). Subclinical first trimester renal abnormalities are associated with preeclampsia in normoalbuminuric women with type 1 diabetes. Diabetes Care, 41(1), 120–127. https://doi.org/10.2337/dc17-1635
Palma, C., Masud, M. K., Guanzon, D., et al. (2025). Rapid and high-sensitivity screening of pregnancy complications by profiling circulating placental extracellular vesicles. Science Advances, 11(9), eadr4074. https://doi.org/10.1126/sciadv.adr4074
Provendier, A., Azria, E., Kayem, G., et al. (2024). The sFlt-1/PlGF ratio for prediction of preeclampsia-related outcomes in women with preexisting diabetes. Reproductive Sciences, 31, 2371–2378. https://doi.org/10.1007/s43032-024-01540-9
Rathnayake, H., Han, L., da Silva Costa, F., et al. (2024). Advancement in predictive biomarkers for gestational diabetes mellitus diagnosis and related outcomes: A scoping review. BMJ Open, 14(12), e089937. https://doi.org/10.1136/bmjopen-2024-089937
Starodubtseva, N., Tokareva, A., Kononikhin, A., et al. (2025). Proteome-based maternal plasma and serum biomarkers for preeclampsia: A systematic review and meta-analysis. Life, 15(5), 776. https://doi.org/10.3390/life15050776
Venkatesh, K. K., Bello, N. A., Tellez Freitas, C. M., et al. (2025). Polygenic risk scores for preeclampsia prediction beyond gold-standard clinical models in multiethnic populations. Journal of the American Heart Association. https://doi.org/10.1161/JAHA.125.046211
Yamazaki, T., Cerdeira, A. S., Teraoka, Y., et al. (2025). Predictive accuracy of sFlt-1/PlGF ratio for preeclampsia and adverse outcomes: Prospective, multicenter observational study in Japan. Hypertension Research, 48, 2548–2557. https://doi.org/10.1038/s41440-025-02282-0
The Implementation Gap
Preeclampsia affects 2–8% of pregnancies globally (70,000 maternal deaths annually), while gestational diabetes mellitus (GDM) complicates 15–20% of pregnancies. Current prediction relies on clinical risk factors achieving only 30–50% sensitivity. Traditional biomarkers (sFlt-1/PlGF) perform well in late pregnancy but lack first-trimester predictive power when intervention is most effective. Breakthrough 2024–2025 validation studies demonstrate cell-free DNA predicts preterm preeclampsia with 81% sensitivity at ≤16 weeks (Adil et al., 2025), cell-free RNA distinguishes disease subtypes 18 weeks before diagnosis (Castillo-Marco et al., 2025), and placental extracellular vesicles identify GDM with 95% sensitivity (Palma et al., 2025). The implementation gap persists: multi-ancestry validation incomplete, clinical workflows undefined, EHR integration pathways unclear, and cost/reimbursement structures unresolved. This brief synthesizes T2 validation evidence and defines T3 implementation readiness criteria.
Evidence for Implementation Readiness
Validated Predictive Models with Clinical Performance
According to PubMed, the PEARL (Preeclampsia Early Assessment of Risk from Liquid biopsy) framework, validated across 1,854 samples at University of Washington/Fred Hutchinson, uses tissue-specific nucleosome profiling from standard NIPT blood draws to predict preterm preeclampsia at ≤16 weeks gestation—12–20 weeks before symptom onset (Adil et al., 2025). Internal validation achieved AUC 0.85 (95% CI: 0.80–0.90) with 81% sensitivity at 80% specificity. External validation confirmed generalizability: AUC 0.84 (0.79–0.89). Early-onset PE (<34 weeks) showed exceptional discrimination: AUC 0.92, 94% sensitivity. Mechanistic signature: decreased placental cfDNA + increased endothelial cfDNA indicates subclinical placental-maternal interface dysfunction.
Spain's PREMOM cfRNA study (NCT04990141), enrolling 9,586 women across 14 hospitals, demonstrated cfRNA enables preeclampsia subtype discrimination months before diagnosis (Castillo-Marco et al., 2025). Early-onset PE (EOPE) prediction at first trimester: AUC 0.88 (internal), 0.87 (external), with 18-week mean lead time. Late-onset PE (LOPE) at second trimester: AUC 0.90 (internal), 0.77 (external), 14.9-week lead time. EOPE signatures reflect decidualization resistance (CBR3, MMP7); LOPE reflects maternal metabolic pathways—enabling personalized intervention (aspirin for EOPE, metabolic management for LOPE).
Table 1. Multimodal Biomarker Platform Performance Metrics
Biomarker Platform | AUC | Sensitivity | Specificity | Lead Time | Key Advantage |
|---|---|---|---|---|---|
cfDNA (PEARL) | 0.85 (0.80–0.90) | 81% | 80% | 12–20 weeks | Bundles with existing NIPT; incremental cost $50–100 |
cfRNA EOPE (T1) | 0.88 (internal) | — | — | 18 weeks | Subtype discrimination enables personalized intervention |
cfRNA LOPE (T2) | 0.90 (internal) | — | — | 14.9 weeks | Maternal metabolic pathway targeting |
Placental EVs (GDM) | — | 95% | 100% | Before 18 weeks | Point-of-care platform; 100% PPV |
sFlt-1/PlGF (Japan) | — | 81.3% | 90% | 1–4 weeks | FDA-cleared; insurance-covered; NPV 98.7% |
Japan's prospective multicenter trial across primary/secondary/tertiary care facilities (n=303) validated sFlt-1/PlGF clinical utility at cut-off 38: NPV 98.7% (95% CI: 96.2–99.3) for ruling out PE within 1 week, PPV 73.7% for adverse outcomes within 4 weeks (Yamazaki et al., 2025). Primary care PPV reached 83.3%, demonstrating usability across all care levels. FDA clearances: Thermo Fisher PreClara (May 2023), Roche Elecsys (February 2025). Japan national insurance coverage since July 2021.
Placental extracellular vesicle nanoplatform for first-trimester GDM prediction achieved 95% sensitivity, 100% specificity, 100% PPV at 11–13 weeks (n=201 women) using CD9+/PLAP+ biomarkers on point-of-care glass strip readout (Palma et al., 2025). Standard fasting glucose achieves only AUC 0.56. Exceptional metrics require independent validation in larger, diverse populations before clinical deployment.
Real-World Implementation and Cost-Effectiveness
PRAECIS US multicenter study (n=1,014; 18 centers; 31% Black, 16% Hispanic) demonstrated sFlt-1/PlGF maintains AUC 0.92 versus 0.75 for standard clinical assessment across racial/ethnic groups. Cost-effectiveness analyses show consistent savings: $10,595/patient (USA), £344/patient (UK), ¥16,373/patient (Japan). PEARL cfDNA projected cost-effective when bundled with NIPT infrastructure—incremental cost $50–100 versus standalone $300–800.
Health Equity Performance Gaps
Critical disparity: Polygenic risk scores show no predictive improvement in women of African ancestry despite improving European-ancestry prediction (Venkatesh et al., 2025). Black women experience 12.4% PE prevalence (versus 7.1% White) and 3.7× increased in-hospital mortality. GDM biomarker scoping review (137 studies, 278 biomarkers) found only 2 studies performed external validation—validated biomarkers remain "scarce and not ready for clinical use" (Rathnayake et al., 2024). PEARL cfDNA and Spain cfRNA require race/ethnicity-stratified validation before broad implementation.
Implementation Solution
Integrated Risk Stratification Algorithm
Sequential biomarker deployment matched to gestational age optimizes sensitivity while controlling costs. The integrated prediction model combines cfDNA/cfRNA for early risk stratification with sFlt-1/PlGF for clinical triage:
First Trimester (9–16 weeks) — Early Risk Stratification:
PEARL model achieves AUC 0.85 when integrating tissue-specific nucleosome patterns with clinical variables. Coefficients (β) derived from XGBoost machine learning. Incremental cost: $50–100 when bundled with existing NIPT infrastructure.
Second Trimester (18–28 weeks) — Subtype Discrimination:
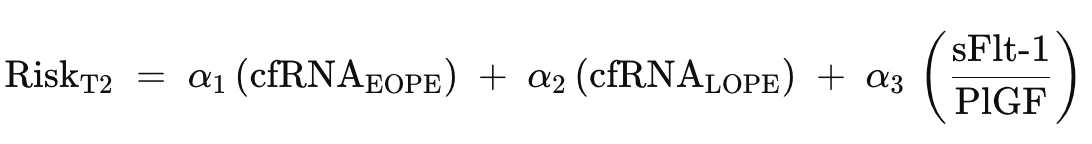
cfRNA enables personalized intervention: EOPE → aspirin + intensive monitoring; LOPE → metabolic management. sFlt-1/PlGF provides established decision support for high-risk patients.
Third Trimester (>28 weeks) — Short-Term Triage:

Clinical Workflow Integration
Deploy via Epic Genomics Module or Cerner Millennium. cfDNA results integrate through existing NIPT reporting infrastructure. sFlt-1/PlGF orders via CPOE with automated Best Practice Alerts for suspected PE. Algorithm-generated risk scores display on chart SnapShot with trimester-specific intervention recommendations.
Table 2. EHR Implementation Timeline
Gestational Age | Biomarker | Action | EHR Integration |
|---|---|---|---|
9–16 weeks | cfDNA + NIPT | Risk stratification: Low/Int/High | Epic Genomics Module auto-populate |
18–28 weeks | cfRNA OR sFlt-1/PlGF (if high-risk) | EOPE: aspirin; LOPE: metabolic mgmt | Best Practice Alert triggers |
>28 weeks | sFlt-1/PlGF (suspected cases) | Discharge (<38) vs Transfer (>85) | Ob Triage decision support |
11–13 weeks (GDM) | Placental EV (if available) | Early lifestyle intervention | Point-of-care result → care plan |
Figure 1: Multimodal Biomarker Clinical Pathway
┌─────────────────────────────────────────────────────────────────────────────┐ │ PREECLAMPSIA & GDM MULTIMODAL BIOMARKER WORKFLOW │ ├─────────────────────────────────────────────────────────────────────────────┤ │ │ │ FIRST TRIMESTER (9-16 weeks) SECOND TRIMESTER (18-28 weeks) │ │ ┌──────────────────┐ ┌───────────────────┐ │ │ │ cfDNA + NIPT │──HIGH RISK──▶ │ cfRNA Subtype OR │ │ │ │ • Nucleosome │ │ sFlt-1/PlGF │ │ │ │ • AUC 0.85 │ │ • EOPE: Aspirin │ │ │ │ • Cost: +$50-100 │ │ • LOPE: Metabolic │ │ │ └────────┬─────────┘ └─────────┬─────────┘ │ │ │ │ │ │ ▼ ▼ │ │ ┌──────────────────┐ ┌───────────────────┐ │ │ │ RISK TIERS │ │ INTERVENTION │ │ │ │ Low: Standard │ │ EOPE → Aspirin + │ │ │ │ Int: Monitor │ │ Monitor │ │ │ │ High → Advanced │ │ LOPE → BP control │ │ │ └──────────────────┘ └───────────────────┘ │ │ │ │ GDM PATHWAY (11-13 weeks) THIRD TRIMESTER (>28 weeks) │ │ ┌──────────────────┐ ┌───────────────────┐ │ │ │ Placental EVs │ │ sFlt-1/PlGF TRIAGE│ │ │ │ • 95% sens │ │ <38 → Discharge │ │ │ │ • 100% spec │ │ (NPV 98.7%) │ │ │ │ • POC platform │ │ >85 → Transfer │ │ │ └────────┬─────────┘ │ (PPV 77.8%) │ │ │ │ └───────────────────┘ │ │ ▼ │ │ ┌──────────────────┐ ┌───────────────────┐ │ │ │ EARLY LIFESTYLE │ │ OUTCOMES │ │ │ │ • Diet + Exercise│ │ • Maternal safety │ │ │ │ • Before OGTT │ │ • Cost savings │ │ │ └──────────────────┘ └───────────────────┘ │ │ │ │ ───────────────────────────────────────────────────────────────────────── │ │ INPUTS: cfDNA nucleosomes • cfRNA transcripts • sFlt-1/PlGF • Placental EVs│ │ METHODS: XGBoost ML • Tissue-specific profiling • Immunoassays │ │ OUTPUT: Risk Score (0-100) + Trimester-specific intervention pathway │ └─────────────────────────────────────────────────────────────────────────────┘
References
Adil, M., Kolarova, T. R., Doebley, A. L., Ha, G., et al. (2025). Preeclampsia risk prediction from prenatal cell-free DNA screening. Nature Medicine, 31(4), 1312–1318. https://doi.org/10.1038/s41591-025-03509-w
Andresen, I. J., Romero, R., Kalapotharakos, G., et al. (2025). Large-scale proteomics reveals new candidate biomarkers for late-onset preeclampsia. Hypertension. https://doi.org/10.1161/HYPERTENSIONAHA.125.25189
Castillo-Marco, N., Cordero, T., Igual, M., et al. (2025). Maternal plasma cell-free RNA as a predictor of early and late-onset preeclampsia throughout pregnancy. Nature Communications, 16, 9208. https://doi.org/10.1038/s41467-025-64215-2
Kelly, C. B., Hookham, M. B., Yu, J. Y., et al. (2018). Subclinical first trimester renal abnormalities are associated with preeclampsia in normoalbuminuric women with type 1 diabetes. Diabetes Care, 41(1), 120–127. https://doi.org/10.2337/dc17-1635
Palma, C., Masud, M. K., Guanzon, D., et al. (2025). Rapid and high-sensitivity screening of pregnancy complications by profiling circulating placental extracellular vesicles. Science Advances, 11(9), eadr4074. https://doi.org/10.1126/sciadv.adr4074
Provendier, A., Azria, E., Kayem, G., et al. (2024). The sFlt-1/PlGF ratio for prediction of preeclampsia-related outcomes in women with preexisting diabetes. Reproductive Sciences, 31, 2371–2378. https://doi.org/10.1007/s43032-024-01540-9
Rathnayake, H., Han, L., da Silva Costa, F., et al. (2024). Advancement in predictive biomarkers for gestational diabetes mellitus diagnosis and related outcomes: A scoping review. BMJ Open, 14(12), e089937. https://doi.org/10.1136/bmjopen-2024-089937
Starodubtseva, N., Tokareva, A., Kononikhin, A., et al. (2025). Proteome-based maternal plasma and serum biomarkers for preeclampsia: A systematic review and meta-analysis. Life, 15(5), 776. https://doi.org/10.3390/life15050776
Venkatesh, K. K., Bello, N. A., Tellez Freitas, C. M., et al. (2025). Polygenic risk scores for preeclampsia prediction beyond gold-standard clinical models in multiethnic populations. Journal of the American Heart Association. https://doi.org/10.1161/JAHA.125.046211
Yamazaki, T., Cerdeira, A. S., Teraoka, Y., et al. (2025). Predictive accuracy of sFlt-1/PlGF ratio for preeclampsia and adverse outcomes: Prospective, multicenter observational study in Japan. Hypertension Research, 48, 2548–2557. https://doi.org/10.1038/s41440-025-02282-0
Turn evidence into everyday care.
No spam, unsubscribe anytime.